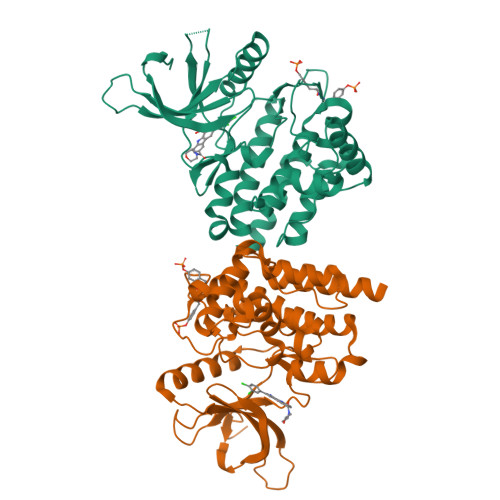
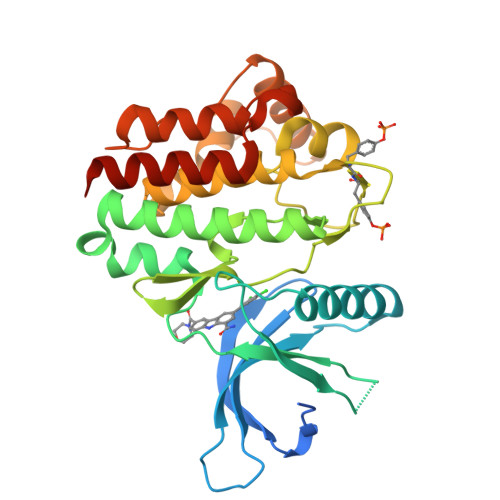
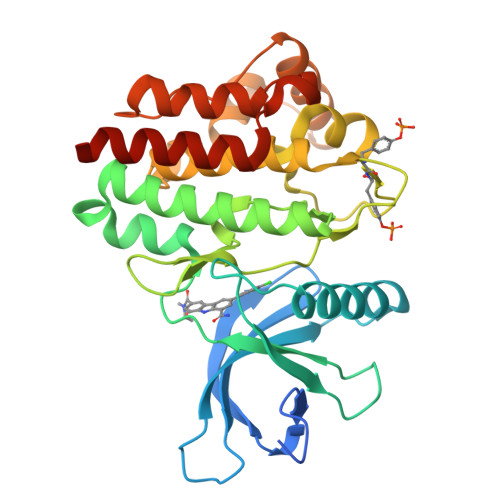
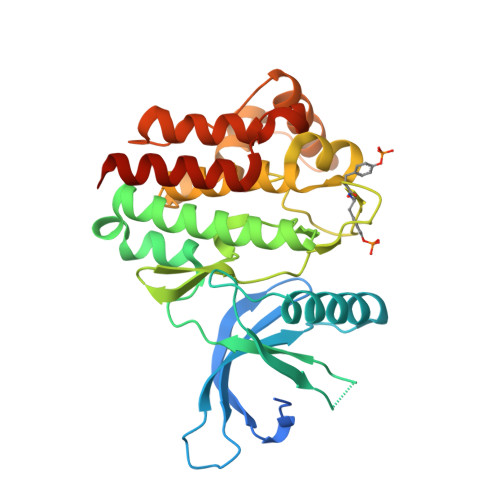
9H-Carbazole-1-carboxamides as potent and selective JAK2 inhibitors.
Zimmermann, K., Sang, X., Mastalerz, H.A., Johnson, W.L., Zhang, G., Liu, Q., Batt, D., Lombardo, L.J., Vyas, D., Trainor, G.L., Tokarski, J.S., Lorenzi, M.V., You, D., Gottardis, M.M., Lippy, J., Khan, J., Sack, J.S., Purandare, A.V.(2015) Bioorg Med Chem Lett 25: 2809-2812
- PubMed: 25987372
- DOI: https://doi.org/10.1016/j.bmcl.2015.04.101
- Primary Citation of Related Structures:
4ZIM - PubMed Abstract:
The discovery, synthesis, and characterization of 9H-carbazole-1-carboxamides as potent and selective ATP-competitive inhibitors of Janus kinase 2 (JAK2) are discussed. Optimization for JAK family selectivity led to compounds 14 and 21, with greater than 45-fold selectivity for JAK2 over all other members of the JAK kinase family.
Organizational Affiliation:
Bristol-Myers Squibb Co., 5 Research Parkway, Wallingford, CT 06492-1951, USA. Electronic address: Kurt.Zimmermann@bms.com.